Disease Diagnosis with OCTA Retinal Scans
An Artificial Intelligence & Data Engineering Design Project
An Artificial Intelligence & Data Engineering Design Project
This project focuses on the automated classification of retinal diseases by leveraging the rich, multi-modal information from Optical Coherence Tomography (OCT) and OCT Angiography (OCTA) scans. While the human eye offers a window to systemic health, diagnosing conditions like Age-related Macular Degeneration (AMD), Diabetic Retinopathy (DR), and even Alzheimer's Disease (AD) from retinal scans is a complex task.
The core contribution is the development of MultiModalNet, a novel dual-branch, hybrid deep learning architecture. This system is designed to overcome the limitations of single-modality models by synergistically processing two complementary data types:
By fusing features from both a Convolutional Neural Network (CNN) and a Vision Transformer (ViT), MultiModalNet provides a more robust and accurate diagnosis, demonstrating a significant performance increase over baseline approaches.
Our model processes two types of 3D retinal scans: OCT for structural detail and OCTA for vascular mapping. To make this complex data usable for 2D deep learning models, we extract specific 2D representations: B-scans from OCT and en-face projection maps from OCTA.
An OCT scan captures a 3D volume of the retina, composed of hundreds of sequential cross-sectional images (B-scans). This provides a detailed structural map of retinal layers, revealing depth and thickness.
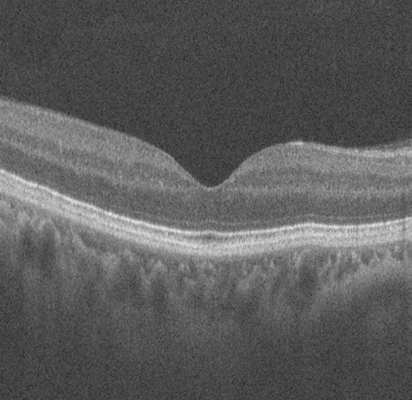
A single B-scan is a cross-sectional slice from the 3D OCT volume. We select representative slices to feed into our model, providing critical information about retinal layer integrity for disease detection.
Similarly, an OCTA scan captures a 3D volume but specifically maps blood flow. This allows for the visualization of the complex, multi-layered vascular network within the retina.
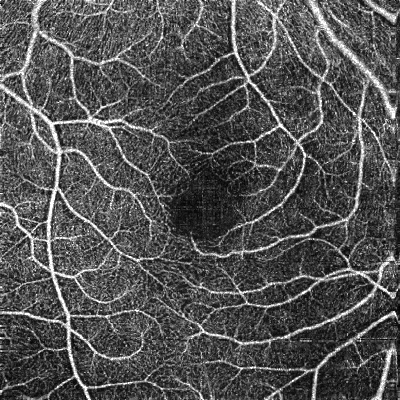
To get a 2D summary of the vasculature, the 3D OCTA volume is "flattened" or projected into an en-face map. This image highlights blood vessel patterns, which is ideal for analysis with CNNs.

MultiModalNet is a dual-branch hybrid deep learning architecture for multi-modal retinal disease classification. It processes two complementary data types in parallel: OCTA projection maps (2D en-face images summarizing vascular patterns, stacked as multi-channel inputs) and B-scan slices (cross-sectional images from 3D OCT volumes, combined to represent retinal structure).
Each modality is handled by a specialized encoder—a ResNet101 CNN for projection maps and a ViT-Large Vision Transformer for B-scans. The extracted features are then fused and passed to a classification layer. This approach combines the local feature extraction strength of CNNs with the global context understanding of Transformers, leading to superior accuracy.
The system supports various models, including the proposed MultiModalNet, ResNet50/101, ViT-Large, ConvNeXt, Masked Autoencoders, RETFound, and TransPro for synthetic data generation. Developed in Python with PyTorch, it employs key techniques like multi-modal learning, feature fusion, transfer learning, self-supervised pre-training, and Focal Loss to handle class imbalance, utilizing the OCTA-500 and UK Biobank datasets.
Standard preprocessing, including resizing, normalization, and augmentation, is applied. The model is trained end-to-end, ensuring robust and generalizable performance across different retinal disease classification tasks.
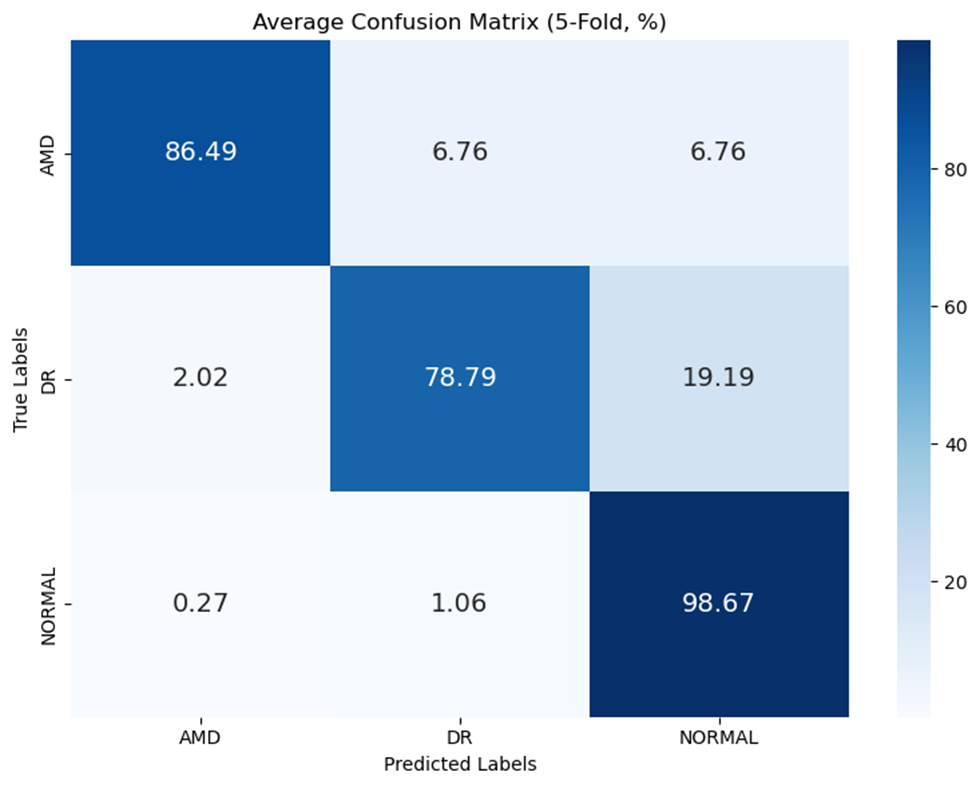
The multi-modal approach demonstrated superior performance compared to single-modality models, validating the core hypothesis of the project. Building upon our previous research, which established a baseline accuracy of 90.41%, this work further refines the model architecture to achieve new state-of-the-art results.
Average Confusion Matrix (5-Fold, %)
Watch the short video below for a high-level overview of the project's goals, methods, and key findings.